ECRcentral: Bringing early-career researchers and funding opportunities together, ISMB/ECCB 2019, Basel. Jul 2019. - [Poster and Talk]
Preprints in the life sciences, Sven Furberg Seminar
in Bioinformatics and Statistical Genomics, Oslo, Norway. Jan 2019. - [Talk]
A dynamic landscape of enhancer clusters during mouse development, CSHL meeting on Epigenetics & Chromatin, New York, USA. Sep 2018. - [Poster]
Towards a format-free submission policy: Let’s put science first, 2nd annual conference of
Digital Life Norway Research School, Stiklestad, Norway. Jun 2018. - [Talk]
JASPAR: a comprehensive database of transcription factor binding profiles, 11th International
Biocuration Conference, Shanghai. Apr 2018. - [Poster and Talk]
Super-enhancers are transcriptionally more active and cell type-specific than stretch enhancers, Genomic Regulation 2018, Haute-Nendaz, Switzerland. Mar 2018. - [Poster]
A novel web framework and RESTful API for the JASPAR database of transcription factor
binding profiles, 3rd annual conference of NORBIS, Tromsø, Norway. Nov 2017. - [Poster]
JASPAR RESTful API: accessing JASPAR data from any programming languages, GREEKC
Training on Knowledge Commons and Tools interoperability, Lisbon, Portugal. Oct 2017. - [Talk]
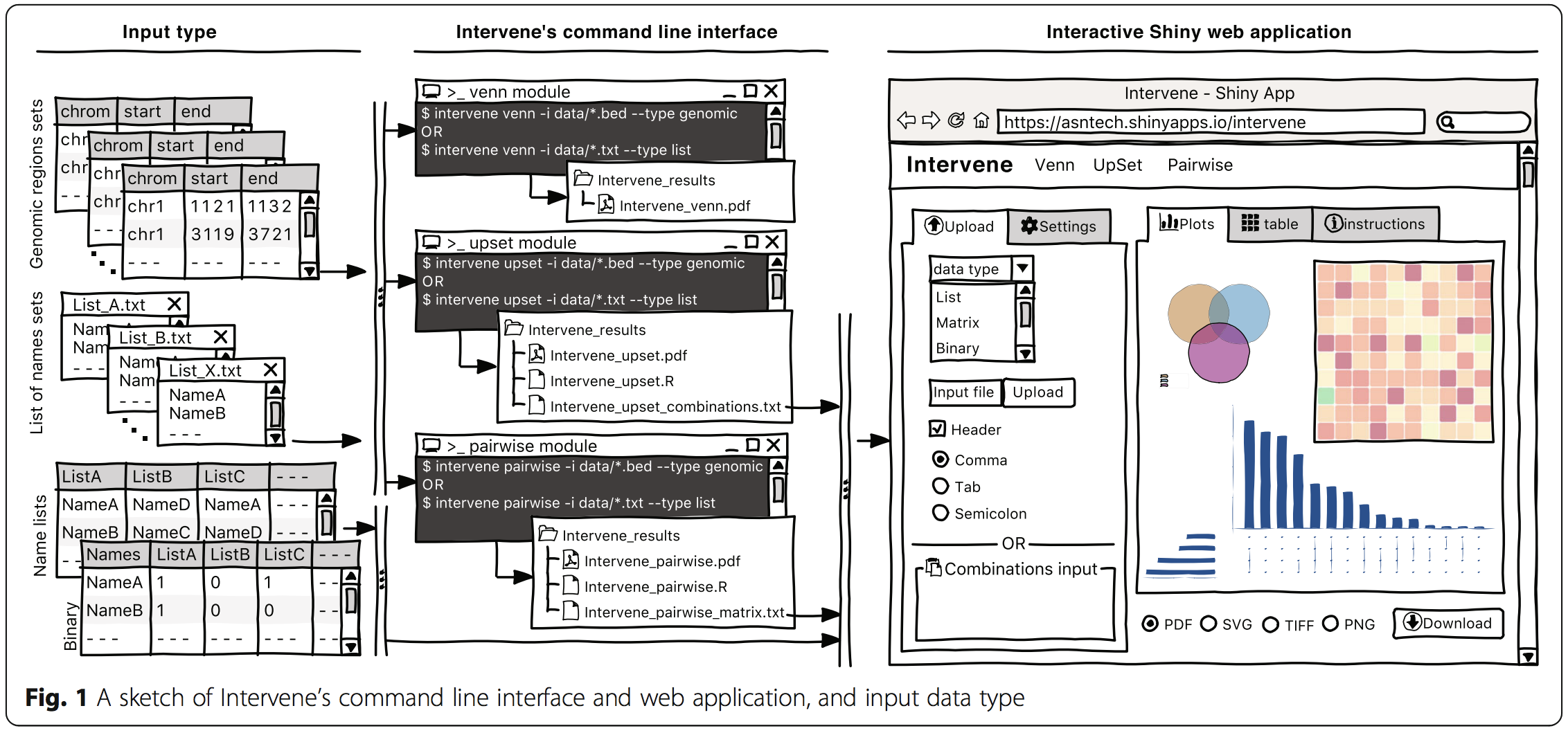
Intervene: a tool for intersection and visualization of multiple gene or genomic region sets,
ISCB Student Council Symposium 2017, Prague, Czech Republic. Jul 2017. - [Talk]
Intervene: a tool for intersection and visualization of multiple gene or genomic region sets,
ISMB/ECCB 2017, Prague, Czech Republic. Jul 2017. - [Poster]
Integrative analysis reveals significant differences between super and stretch enhancers, Gene
Expression Data Analysis , Mainz, Germany. Nov 2016. - [Poster and Talk]
Comprehensive analysis, prediction, and annotation of super-enhancers, Sven Furberg Seminar
in Bioinformatics and Statistical Genomics, Oslo, Norway. Apr 2017. - [Talk]
Computational prediction, characterization & annotation of super-enhancers & analysis of their
key features, CSHA - Chromatin, Epigenetics & Transcription, Suzhou, China. May 2016. - [Poster]
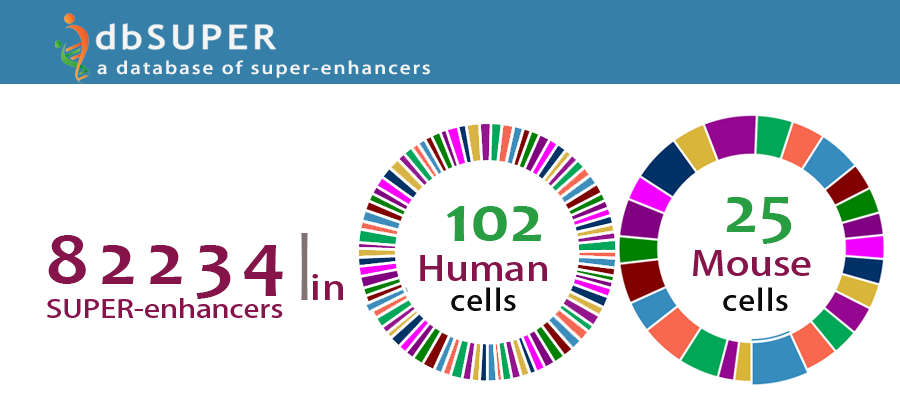
dbSUPER - an extensive and interactive database of super-enhancers, CSHA/AACR Meeting -
Big Data, Comp. & Sys. Bio. in Cancer, Suzhou, China. Dec 2015. - [Poster]
Super-enhancer prediction from epigenetic signatures and sequence motif data, ISMB/ECCB
2015, Dublin, Ireland. Jul 2015. - [Poster]
An extensive and interactive database of super-enhancers – dbSUPER, ISCB Student Council
Symposium 2015, Dublin, Ireland, Jul 2015. - [Poster]
An extensive and interactive database of super-enhancers – dbSUPER, 8th International
Biocuration Conference (IBC), Beijing. Apr 2015. - [Poster]
Integration of ChIP-seq and machine learning reveals super-enhancers and features of
biological significance, Synthetic Biology Workshops, Tsinghua University. May 2014. - [Talk]